Import the dataset DATI OBESI https://docs.wixstatic.com/ugd/288b42_8ab73b0b44254f649bc9bf1f75a90780.xls?dn=DATI_OBESI.xls in R, check and, if necessary, convert the variable classes, display the number of samples and variables, and perform a descriptive statistics analysis of the entire dataframe.
Different procedures can be used to load data files into R. It depends on the file type. The most common and straightforward file types are ".txt" and ".csv."
In ".txt" and ".csv" files, the columns are separated by symbols such as tabs or commas. We can always convert a file to one of these types by saving it in tab-delimited text format by selecting "Save As" → "Text (tab-delimited) .txt." Alternatively, save as "CSV (comma-separated)." If we open the file in Notepad or any text editor, we will see data separated by tabs or commas.
If we choose to convert the original file to a .txt or .csv file, we can load it using the read.table() function.
The syntax to import the data will be:
DATI_OBESI<- read.table("/path/to/file/File_name.txt", header=TRUE, sep="\t") DATI_OBESI<- read.table("/path/to/file/File_name.csv", header=TRUE, sep=",")To find the file path, right-click the data file, select Properties, and copy the path.
Make sure the slashes are "/" instead of "".
header=TRUE indicates that the file has a header row (with variable names).
sep="\t" specifies that columns are separated by tabs.
For CSV files, the separator is sep=",".
If your computer’s settings are in Italian, it’s possible the CSV file uses semicolons instead of commas. In that case, set sep=";".
Alternatively, you can install a package that allows you to load .xls files directly, called xlsx. After installing, load it into R to access its functions, including read.xlsx().
install.packages("xlsx")
library(xlsx)
DATI_OBESI <-read.xlsx("C:/Users/GENTILINI/Desktop/DATI_OBESI.xls", sheetName = "DATI_OBESI")Display the file structure and variable attributes:
str(DATI_OBESI)Reclassify categorical variables that R has misinterpreted as numeric:
DATI_OBESI$IID<-as.factor(DATI_OBESI$IID)
DATI_OBESI$OBESITA_GRAVE<-as.factor(DATI_OBESI$OBESITA_GRAVE)
DATI_OBESI$sesso<-as.factor(DATI_OBESI$sesso)
DATI_OBESI$diabete<-as.factor(DATI_OBESI$diabete)
DATI_OBESI$tipo_diabete<-as.factor(DATI_OBESI$tipo_diabete)
DATI_OBESI$ipertensione<-as.factor(DATI_OBESI$ipertensione)
DATI_OBESI$OB_EDONICI<-as.factor(DATI_OBESI$OB_EDONICI)
DATI_OBESI$INFARTO<-as.factor(DATI_OBESI$INFARTO)Display the number of samples and variables in the file:
dim(DATI_OBESI)Impute missing data in numeric variables using the mean.
Install and load the mice package:
install.packages("mice")
library(mice)Impute the dataset, specifying that the continuous variables are in columns 4 to 41.
DATI_IMPUT<-mice(DATI_OBESI[,4:41],m=1,method ="pmm",seed=500)
summary(DATI_IMPUT)
completedData<- complete(DATI_IMPUT,1)Display the imputed dataset containing the continuous variables.
Display the distribution of the variables “s col” and “s hdl”:
densityplot(completedData$s_col)
densityplot(completedData$s_hdl)Using an appropriate graph, display the median, quartiles, and any outliers of the variables "peso" (weight), "alt" (height), "BMI," and "fian" (hip circumference). Ensure that for these four variables, comparisons are made between males and females:
boxplot(completedData$peso~DATI_OBESI$sesso)
boxplot(completedData$bmi~DATI_OBESI$sesso)
boxplot(completedData$alt~DATI_OBESI$sesso)
boxplot(completedData$fian~DATI_OBESI$sesso)
Using an appropriate graph, display any relationship between "pasis" (systolic pressure) and "padia" (diastolic pressure):
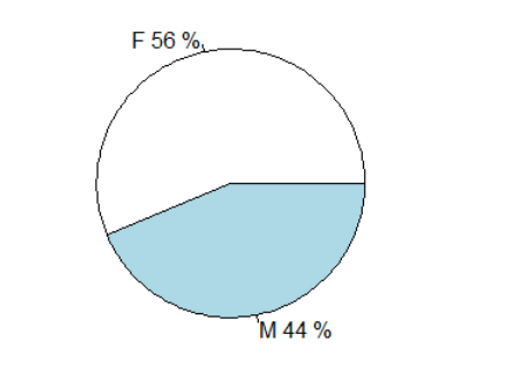
scatter.smooth(completedData$pasis~completedData$padia, xlab = "PRESSIONE DIASTOLICA", ylab = "PRESSIONE SISTOLICA")How many males and females are there? Display the proportions using the most appropriate graph.
Create a pie chart that also shows percentages:
percentuali <-round(table(DATI_OBESI$sesso)/sum(table(DATI_OBESI$sesso))*100)
labels<-paste(levels(DATI_OBESI$sesso), percentuali,"%", sep=" ")
pie(table(DATI_OBESI$sesso),labels)How many individuals are severely obese (1= Severely Obese, 2= Not Severely Obese)? How many of them are male?
table(DATI_OBESI$OBESITA_GRAVE) table(DATI_OBESI$OBESITA_GRAVE,DATI_OBESI$sesso)The severely obese individuals are 740, with 290 being male.
Display the mean values of "ins" (Insulin) using a barplot, divided into severely obese and non-severely obese groups:
barplot(tapply(completedData$s_ldl,DATI_OBESI$OBESITA_GRAVE,mean))How many individuals are diabetic, hypertensive, and have had a heart attack? (1=yes, 2=no)
Replace the "0,1" levels of the categorical variables with words that clearly indicate the levels of the three categorical variables:
levels(DATI_OBESI$INFARTO)<-c("INFARTOSI","INFARTONO")
levels(DATI_OBESI$diabete)<-c("DIABETESI","DIABETENO")
levels(DATI_OBESI$ipertensione)<-c("IPERTSI","IPERTINO")Use the table command to find the number of diabetic, hypertensive, and heart attack patients:
table(DATI_OBESI$ipertensione,DATI_OBESI$diabete,DATI_OBESI$INFARTO)There are 450 individuals with heart attacks, diabetes, and hypertension.
Select the subjects described in point 9 and display the mean levels of "s ldl" (LDL cholesterol) comparing these means among severely obese and non-severely obese individuals using a barplot.
Using subset, create an object containing only the diabetic, hypertensive, and heart attack subjects:
DIABIPERTINFAR<- subset(DATI_OBESI,DATI_OBESI$diabete=="DIABETESI" & DATI_OBESI$INFARTO=="INFARTOSI" & DATI_OBESI$ipertensione=="IPERTSI")Proceed with the graphical analysis:
barplot(tapply(DIABIPERTINFAR$s_ldl,DIABIPERTINFAR$OBESITA_GRAVE,mean))